Together with Dino, Sam, Zoltan, and Arthur, I recently arxived a first draft published an article on a project that combines two topics — the combination of which I find rather exciting: kernel methods and Hamiltonian Monte Carlo.
Updates.
- January: Presented the poster at MCMSki
- December III: Arthur gave a talk (slides) the scalable Monte Carlo workshop at NIPS 2015
- December II: We had a poster at NIPS
- December I: NIPS camera ready version now on arXiv
- November: Implementation of kernel hmc online
- October: Implementation of gradient estimation online
- September: Accepted at NIPS 2015! (and a name change to come …)
- July: Our reply to Xian’s blog post was also featured. Check out the IPython notebook I wrote for that.
- June 2015: Our draft was discussed on Xian’s Og and caused a lively discussion on the sense and non-sense of pseudo-marginal samplers and regularisation strength of kernel methods.
Here is the abstract.
We propose
KamiltonianKernel Hamiltonian Monte Carlo (KMC), a gradient-free adaptive MCMC algorithm based on Hamiltonian Monte Carlo (HMC). On target densities where classical HMC is not an option due to intractable gradients, KMC adaptively learns the target’s gradient structure by fitting an exponential family model in a Reproducing Kernel Hilbert Space. Computational costs are reduced by two novel efficient approximations to this gradient. While being asymptotically exact, KMC mimics HMC in terms of sampling efficiency, and offers substantial mixing improvements over state-of-the-art gradient free samplers. We support our claims with experimental studies on both toy and real-world applications, including Approximate Bayesian Computation and exact-approximate MCMC.
Motivation: HMC with intractable gradients.
Many recent applications of MCMC focus on models where the target density function is intractable. A very simple example is the context of Pseudo-Marginal MCMC (PM-MCMC), for example in Maurizio’s paper on Bayesian Gaussian Processes classification. In such (simple) models the marginal likelihood $p(\mathbf{y}|\theta )$ is unavailable in closed form, but only can be estimated. Performing Metropolis-Hastings style MCMC on $\hat{p}(\theta |\mathbf{y})$ results in a Markov chain that (remarkably) converges to the true posterior. So far so good. But no gradients.
 Sometimes, people argue that for simple objects as latent Gaussian models, it is possible to side-step the intractable gradients by running an MCMC chain on the joint space $(\mathbf{f},\theta)$ of latent variables and hyper-parameters, which makes gradients available (and also comes with a set of other problems such as high correlations between $\mathbf{f}$ and $\theta$, etc). While I don’t want to get into this here (we doubt existence of a one-fits-all solution), there is yet another case where gradients are unavailable.
Sometimes, people argue that for simple objects as latent Gaussian models, it is possible to side-step the intractable gradients by running an MCMC chain on the joint space $(\mathbf{f},\theta)$ of latent variables and hyper-parameters, which makes gradients available (and also comes with a set of other problems such as high correlations between $\mathbf{f}$ and $\theta$, etc). While I don’t want to get into this here (we doubt existence of a one-fits-all solution), there is yet another case where gradients are unavailable.
Approximate Bayesian Computation is based on the context where the likelihood itself is a black-box, i.e. it can only be simulated from. Imagine a physicist coming to you with three decades of intuition in the form of some Fortran code, which contains bits implemented by people who are not alive anymore — and he wants to do Bayesian inference on the code-parameters via MCMC… Here we have to give up on getting the exact answer, but rather simulate from a biased posterior. And of course, no gradients, no joint distribution.
 State-of-the-art methods on such targets are based on adaptive random walks, as no gradient information is available per-se. The Kameleon (KAMH) improves over other adaptive MCMC methods by constructing locally aligned proposal covariances. Wouldn’t it be cooler to harness the power of HMC?
State-of-the-art methods on such targets are based on adaptive random walks, as no gradient information is available per-se. The Kameleon (KAMH) improves over other adaptive MCMC methods by constructing locally aligned proposal covariances. Wouldn’t it be cooler to harness the power of HMC?
Kamiltonian Monte Carlo starts as a Random Walk Metropolis (RWM) and then smoothly transitions into HMC. It explores the target at least as fast as RWM (we proof that), but improves the mixing in areas where it has been before.
We do this by learning the target gradient structure from the MCMC trajectory in an adaptive MCMC framework — using kernels. Every MCMC iteration, we update our gradient estimator with a decaying probability $a_t$ that ensures that we never stop updating, but update less and less, i.e. $$\sum_{t=1}^\infty \frac{1}{a_t}=\infty\qquad\text{and}\qquad\sum_{t=1} ^\infty \frac{1}{a_t^2}=0.$$ Christian Robert challenged our approach: using non-parametric density estimates for MCMC proposals directly is a bad idea: if certain parts of the space are not explored before adaptation effectively stopped, the sampler will almost never move there. For KMC (and for KAMH too) however, this is not a problem: rather than using density estimators as proposal directly, we use them for proposal construction. This way, these algorithms inherit ergodicity properties from random walks. I coded an example-demo here.
Kernel exponential families as gradient surrogates.
The density (gradient) estimator itself is an infinite dimensional exponential family model. More precisely, we model the un-normalised target log-density $\pi$ as an RKHS function $f\in\mathcal{H}$, i.e. $$\text{const}\times\pi(x)\approx\exp\left(\langle f,k(x,\cdot)\rangle_{{\cal H}}-A(f)\right),$$ which in particular implies $\nabla f\approx\nabla\log\pi.$ Surprisingly, and despite a complicated normaliser $A(f)$, such a model can be consistently fitted by directly minimising the expected $L^2$ distance of model and true gradient, $$J(f)=\frac{1}{2}\int\pi(x)\left\Vert \nabla f(x)-\nabla\log \pi (x)\right\Vert _{2}^{2}dx.$$ The magic word here is score-matching. You could ask: “Why not use kernel density estimation?” The answer: “Because it breaks in more than a few dimensions.” In contrast, we are actually able to make the above gradient estimator work in ~100 dimensions on Laptops.
Two approximations.
The über-cool infinite exponential family estimator, like all kernel methods, doesn’t scale nicely in the number $n$ of data used for estimation — and here neither in the in input space dimension $d$. Matrix inversion with costs $\mathcal{O}(t^3d^3)$ becomes a blocker, in particular as $t$ here is the growing number of MCMC iterations. KMC comes in two variants, which correspond to different approximations to the original kernel exponential family model.
- KMC lite expresses the log-density in a smaller dimensional (yet growing) sub-space, via collapsing all $d$ input space dimensions into one. It takes the dual form $$f(x)=\sum_{i=1}^{n}\alpha_{i}k(x_{i},x),$$ where the $x_i$ are $n$ random sub-samples (just like KAMH) from the Markov chain history. Downside: KMC lite has to be re-estimated whenever the $x_i$ change. Advantage: The estimator’s tails vanish outside the $x_i$, i.e. $\lim_{\|x\|_{2}\to\infty}\|\nabla f(x)\|_{2}=0$, which translates into a geometric ergodicity result as we will see later.
- KMC finite approximates the model as a finite dimensional exponential family in primal form, $$f(x)=\langle\theta,\phi_{x}\rangle_{{\cal H}_{m}}=\theta^{\top}\phi_{x},$$ where $x\in\mathbb{R}^{d}$ is embedded into a finite dimensional feature space $\phi_{x}\in{\cal H}_{m}=\mathbb{R}^{m}.$ While other choices are possible, we use the Randon Kitchen Sink framework: a $m$-dimensional data independent random Fourier basis. Advantage: KMC lite is an efficient on-line estimator that can be updated at constant costs in $t$ — we can fit it on all of the MCMC trajectory. Disadvantage: Its do not decay and our proof for geometric ergodicity of KMC lite does not apply.

Increasing dimensions.
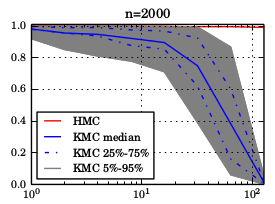
So far, we did not work out how the approximation errors propagate through the kernel exponential family estimator, but we plan to do that at some point. The paper contains an empirical study which shows that the gradients are good enough for HMC up to ~100 dimensions — Under a “Gaussian like smoothness” assumption. The below plots show the acceptance probability along KMC trajectories and quantify how “close” KMC proposals are to HMC proposals.
From RWM to HMC.
Using the well known and (ab)used Banana density as a target, we feed a non-adaptive version of KMC and friends with an increasing number of so-called “oracle” samples (iid from the target), and then quantify how well they mix. While this scenario is totally straw-man, it allows to compare the mixing behaviour of the algorithms after a long burn-in. The below plots show KMC transitioning from a random walk into something close to HMC as the number of “oracle” samples (x-axis) increases.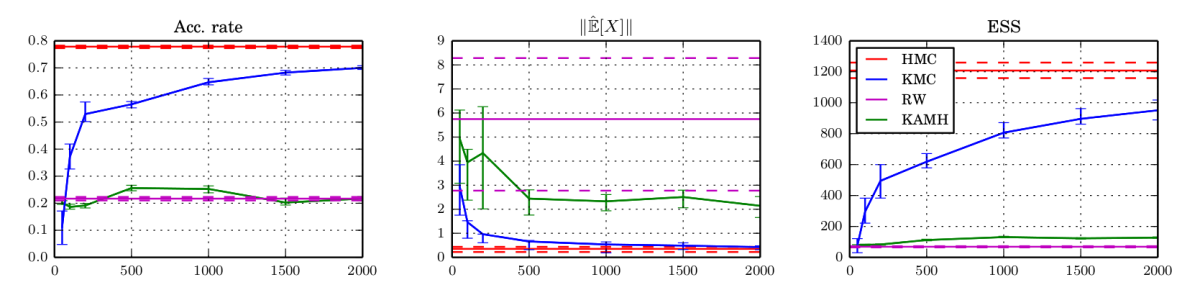 ABC — Reduced simulations and no additional bias.
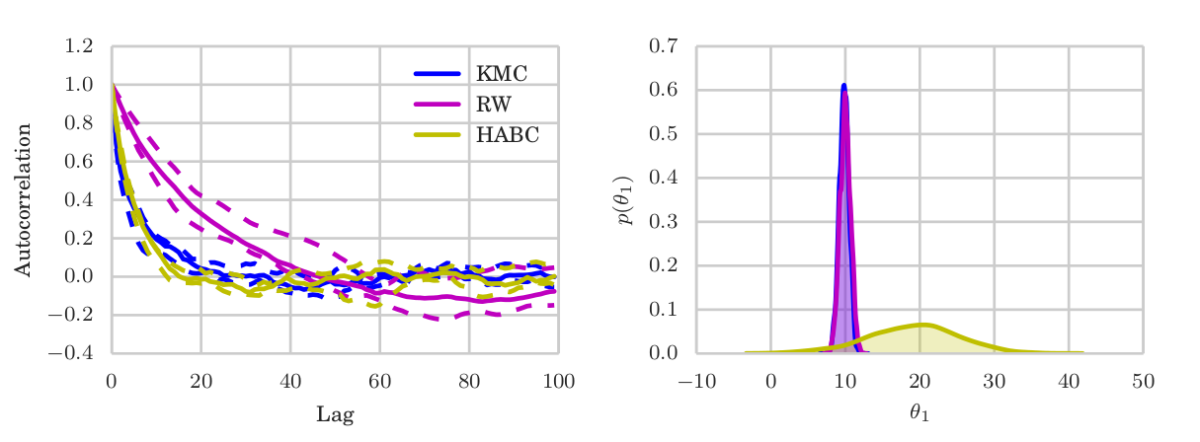
ABC — Reduced simulations and no additional bias.
While there is another example in the paper, I want to show the ABC one here, which I find most interesting. Recall in ABC, the likelihood is not available. People often use synthetic likelihoods that are for example Gaussian, which induces an additional bias (in addition to ABC itself) but might improve statistical efficiency. In an algorithm called Hamiltonian ABC, such a synthetic likelihood is combined with stochastic gradient HMC (SG-HMC) via randomized finite differences, called simultaneous perturbation stochastic approximation (SPSA), which works as follows. To evaluate the gradient at a position $\theta$ in sampling space:
- Generate a random SPSA mask $\Delta$ (set of binary directions) and compute the perturbed $\theta+\Delta$ and $\theta+\Delta$.
- Interpolate linearly between them the perturbations. That is simulate from the ABC likelihood at both points, construct the synthetic likelihood, and use their difference.
- The gradient is a (step-size dependent!) approximation to the unknown gradient of the (biased) synthetic likelihood model.
- Perform a single stochastic HMC leapfrog step (adding friction as described in the SG-HMC paper)
- Iterate for $L$ leapfrog iterations.
What I find slightly irritating is that this algorithm needs to simulate from the ABC likelihood in every leapfrog iteration — and then discards the gained information after a leapfrog step has been taken. How many leapfrog steps are common in HMC? Radford Neal suggests to start with $L\in[100,1000]$. Quite a few ABC simulations come with this! But there are more issues:
- SG-HMC mixing. I found that stochastic gradient HMC mixes very poorly when the gradient noise large. Reducing noise costs even more ABC simulations. Wrongly estimated noise (non-stationary !?!) induces bias due to the “always accept” mentality. Step-size decreasing to account for that further hurts mixing.
- Bias. The synthetic likelihood can fail spectacularly, if the true likelihood is skewed for example.
KMC does not suffer from either of those problems: It keeps on improving its mixing as it sees more samples, while only requiring a single ABC simulation at each MCMC iterations — rather than HABC’s $2L$ (times noise-reduction-repetitions). It targets the true (modulo ABC bias) posterior, while accumulating information of the target (and its gradients) in the form of the MCMC trajectory.
But how do you choose the kernel parameter?
Often, this question is a threat for any kernel-based algorithm. For example, for the KAMH algorithm, it is completely unclear (to us!) how select these parameters in a principled way. For KMC, however, we can simply use cross-validation on the score matching objective function above. In practice, we use a black box optimisation procedure (for example CMA or Bayesian optimisation) to on-line update the kernel hyper-parameters every few MCMC iterations. See an example where our Python code does this here. Just like the updates to the gradient estimator itself, this can be done with a decaying probability to ensure asymptotic correctness.